Regression Modeling Strategies 3 - Logistic Regression
Analyzing Binary and Binomial Outcome Data in Sports Science
Logistic regression, binary outcome, odds ratio, classification, sports analytics, probability modeling, predictive analytics, binomial assumptions.
The level of this lesson is categorized as BRONZE.
- Understanding when and how to apply logistic regression models effectively.
- Identifying binomial random variables and assessing the validity of the binomial assumptions.
- Interpreting estimated coefficients and odds ratios in logistic regression.
In this lesson, we use the HansIll dataset from the speedsR package, an R data package specifically designed for the SPEEDS project. This package provides a collection of sports-specific datasets, streamlining access for analysis and research in the sports science context.
1 Learning Outcomes
By the end of this lesson, you will have developed the ability to:
- Construct Binary Logistic Regression Models: Build logistic regression models in R to analyze binary outcome data.
- Evaluate Model Assumptions: Use diagnostic tools to examine the assumptions of logistic regression models and ensure their validity.
- Interpret Regression Outputs: Interpret the parameters, odds ratios, and test estimates from logistic regression models to draw meaningful insights into relationships between variables.
2 Introduction: Logistic Regression
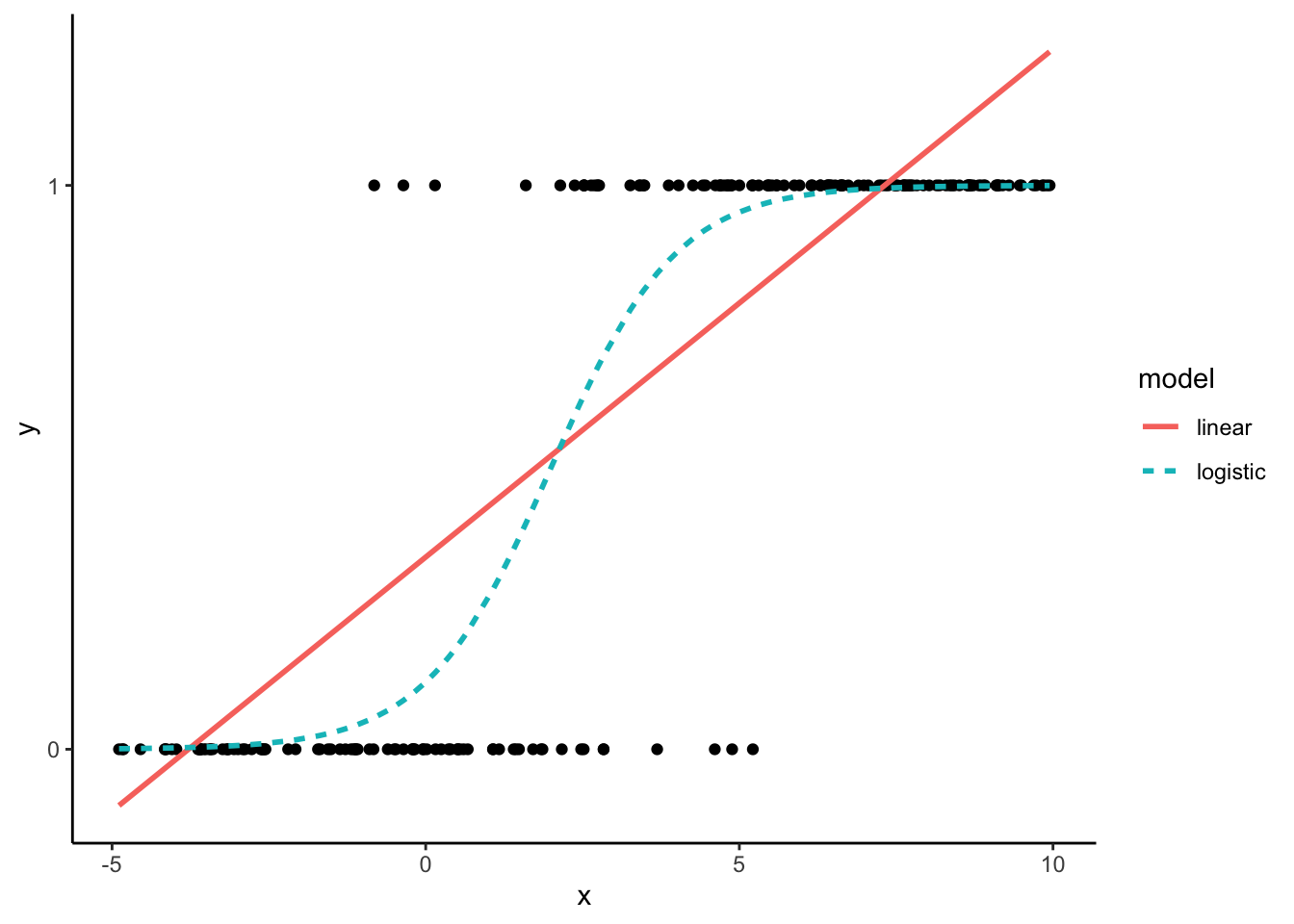
Logistic regression is a versatile and widely used statistical technique designed for modeling relationships between variables, particularly when the dependent variable represents binary outcomes. This methodology is particularly well-suited for scenarios where the outcome of interest is a binary event, such as whether a team wins or loses a match, whether a patient has a disease, or whether a customer makes a purchase. To visualise this, consider the plot below. This plot displays binary data, with responses being either 0’s or 1’s. Two models are fitted to the data (a linear and a logistic regression):
The linear regression model (depicted by the red line) operates under the assumption that the data is continuous, extending beyond the boundaries of 0 and 1. Conversely, the logistic regression model (indicated by the blue line) approaches 0 and 1 but never actually reaches them. This characteristic renders the logistic regression model more suitable for binary data.
The fundamental form of the logistic regression model is expressed as:
\[P(Y=1|X) = \frac{1}{1 + e^{-(\beta_{0} + \beta_{1}X_{1} + \beta_{2}X_{2} + \ldots + \beta_{n}X_{n})}}\]
where \(P(Y=1|X)\) represents the probability of the binary outcome \(Y\) being 1 given the predictors \(X_1, X_2, ..., X_n\), \(\beta_0\) is the intercept, \(\beta_1, \beta_2, ..., \beta_n\) are the coefficients associated with the independent variables \(X_1, X_2, ..., X_n\), and \(e\) denotes the base of the natural logarithm. The objective of logistic regression is to estimate the coefficients that maximize the likelihood of observing the given binary outcome data.
3 Assumptions
Logistic regression adheres to the following statistical assumptions:
- Binary or Binomial Response: The response variable is either binary, representing two possible outcomes (e.g., success or failure, presence or absence), or binomial, representing the count of successes out of a fixed number of trials.
- Independence: Each observation or case in the dataset is assumed to be independent of all other observations. This assumption ensures that there is no systematic relationship between the observations.
- Linearity of Log-Odds: The relationship between the log-odds of the outcome variable and the predictor variable(s) is assumed to be linear. In other words, the logit function (log-odds) of the probability of the event occurring is a linear function of the predictors.
- No Multicollinearity: There should be no multicollinearity among the predictor variables. Multicollinearity occurs when two or more predictor variables are highly correlated, which can lead to unstable estimates of the coefficients.
4 Case Study
4.1 What to expect
In this example we will re-examine the HansIll dataset (see Regression Modeling Strategies 2 - Poisson Regression for more details). We will use the variable Crit1and2 as our dependent variable. Crit1and2 is a binary variable that represents whether participants meet the criteria 1 and 2 of the Jackson’s common cold score (basically, do they show signs of having the common cold).
We will learn how to fit logistic regression models that contain both single and multiple predictors. We will also construct models that include interaction terms. Finally, we will use residual diagnostic tests to inspect the assumptions of our models.
4.2 Data Loading and Cleaning
First, we’ll load the HansIll dataset from the speedsR package into our R environment.
# Load the speedsR package
library(speedsR)
# Access and assign the dataset to a variable
Illness <- HansIll4.3 Initial Exploratory Analyses
As always, it is good practice to explore our data descriptively, before we build our models. Much of this has already been covered in Exploratory Data Analysis (EDA) in Sport and Exercise Science, so we will only provide a short example here.
4.3.1 Data organisation
HansIll is a dataset that contains 39 variables. Some of the more relevant variables (to this exercise) are listed below:
ID: The participant’s ID numbergroup: The testing condition for the participant, 4x(8x40/20s), 4x(12x40/20s) or 4x8mintime: a binary factor to indicate results for either pre or post testsCrit1and2: A binary (true / false) variable that displays whether participant meets criteria 1 AND 2 of the Jackson Common Cold scorevo2_rel: Relative VO2peak (L/min)
Note that for this lesson we will only be using the post-test measurements. As such we should create a filter to only include these measurements (i.e. when time is post). The code below has also renamed the Crit1and2 variable to Cold and changed the labels to ‘Yes’ and ‘No’:
Illness_post <- Illness |>
filter(time == 'post') |>
dplyr::select('group', 'Crit1and2', 'vo2_rel') |>
rename(Cold = Crit1and2) |>
mutate(Cold = factor(case_when(
Cold == TRUE ~ 'Yes',
Cold == FALSE ~ 'No'
))) |>
mutate_if(is.character, as.factor) |>
# Also create a variable that codes Cold into 0s and 1s
mutate(Cold1 = ifelse(Cold == 'Yes', 1, 0))4.3.2 Summary statistics
We can use the skimr package to quickly obtain summary statistics for our chosen variables:
skimr::skim(Illness_post)| Name | Illness_post |
| Number of rows | 25 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| factor | 2 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| group | 0 | 1 | FALSE | 3 | 4x(: 9, 4x(: 8, 4x8: 8 |
| Cold | 0 | 1 | FALSE | 2 | No: 17, Yes: 8 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| vo2_rel | 0 | 1 | 65.50 | 6.81 | 52.1 | 61.2 | 65.4 | 69.4 | 78.7 | ▂▇▇▅▃ |
| Cold1 | 0 | 1 | 0.32 | 0.48 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | ▇▁▁▁▃ |
We can use the table and prop.table functions to explore the Cold variable further:
table(Illness_post$Cold)
No Yes
17 8 prop.table(mosaic::tally(~Illness_post$Cold))Illness_post$Cold
No Yes
0.68 0.32 Next, let us examine this Cold proportion over the group factor:
Illness_post |>
group_by(group) |>
count(Cold) |>
mutate(prop = n/sum(n)) |>
ggplot(aes(group, prop)) +
geom_col(aes(fill = Cold)) +
geom_text(aes(label = scales::percent(prop),
y = prop,
group = Cold),
position = position_stack(vjust = 0.5))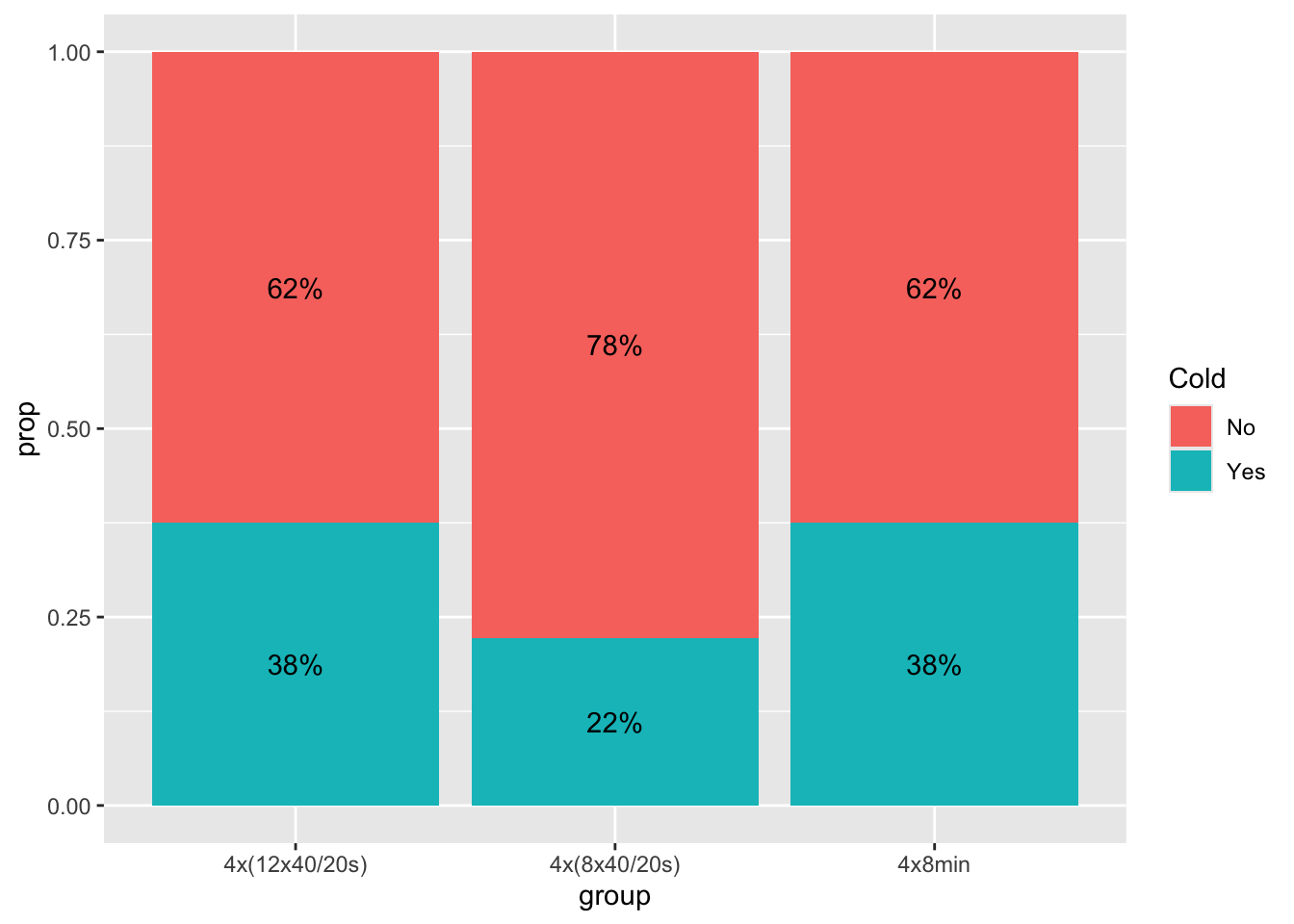
If we consider including a continuous term in our model (in this case: vo2_rel), the logit should be linearly related. We can investigate this assumption by constructing an empirical logit plot. In order to calculate empirical logits, we first divide our data by group. Within each group, we divide vo2_rel into subsets of equal sizes. Given the small size of this data set, we will use three groups:
Illness_post <- Illness_post %>%
group_by(group) %>%
mutate(cuts = cut_number(vo2_rel,3)) |>
ungroup()Within each group, we calculate the proportion, \(\hat{p}\) that reported meeting the criteria for the common cold, and then the empirical log odds, \(log(\frac{\hat{p}}{1-\hat{p}})\), that a person meets this criteria:
emplogit1 <- Illness_post %>%
group_by(group, cuts) %>%
summarise(prop.Cold = mean(Cold1),
n = n(),
midpoint = median(vo2_rel)) %>%
mutate(prop.Cold = ifelse(prop.Cold==0, .01, prop.Cold),
emplogit = log(prop.Cold / (1 - prop.Cold)))Finally, we can plot this data:
ggplot(emplogit1, aes(x = midpoint, y = emplogit, color = group)) +
geom_smooth(method = "lm", se=F)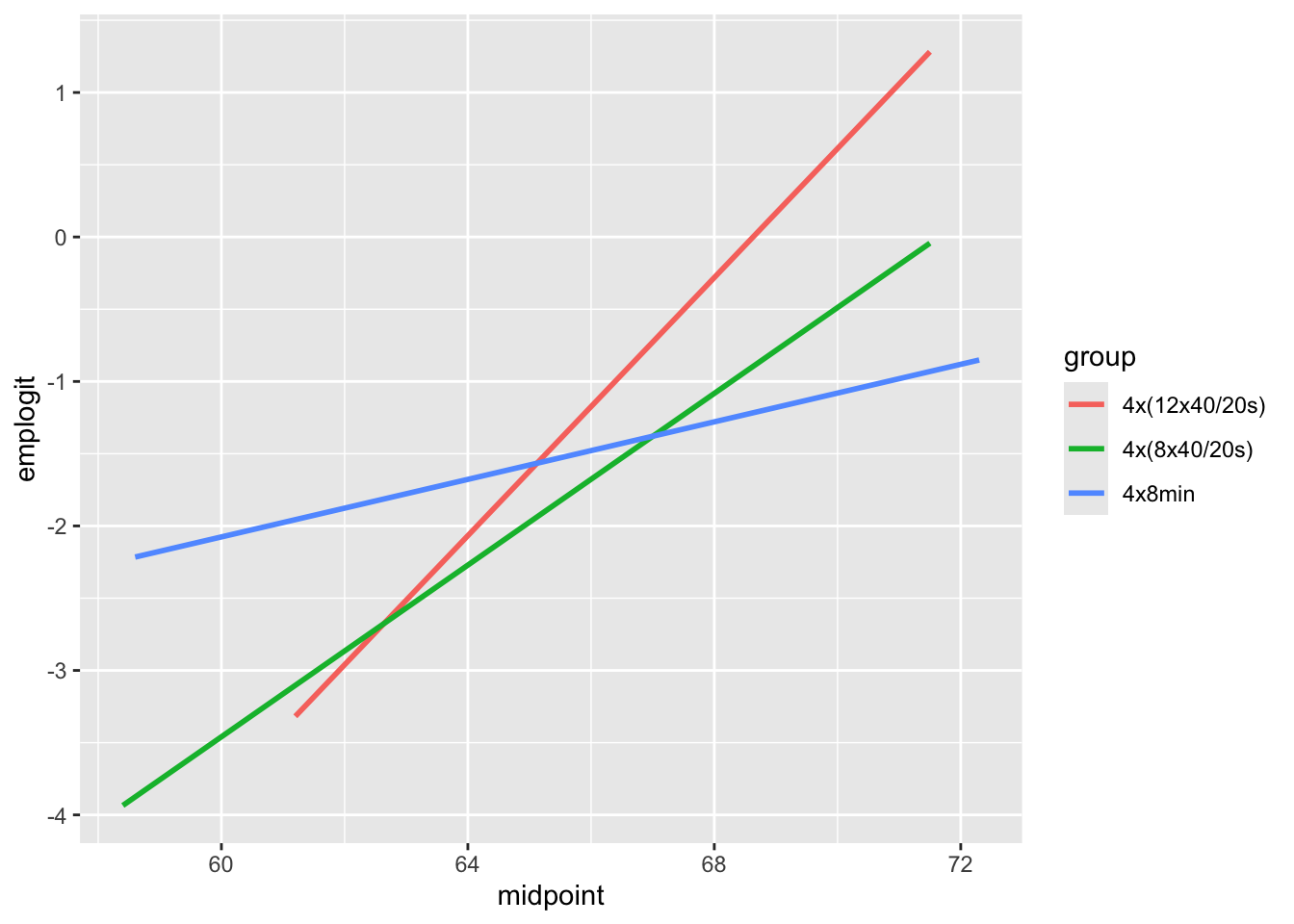
From this plot we can see that all three groups exhibit an increasing linear trend on the logit scale indicating that increasing vo2_rel is associated with a higher chance of having the cold and that modeling log odds as a linear function of vo2_rel is reasonable. The slopes between the three groups are not similar, so we should consider an interaction term between vo2_rel and group in the model.
4.4 Estimation and Inference
4.4.1 Continuous predictor
To begin, we will consider a simple model with only one predictor: vo2_rel. In R, we can use the glm() function, specifying the formula, family and data. For logistic regression, we will specify the family to be binomial.
model1 <- glm(Cold ~ vo2_rel,
family = binomial,
data = Illness_post)We can then use the coef() function to obtain the coefficients:
coef(model1)(Intercept) vo2_rel
-15.4722880 0.2202615 From this output, we can see that the intercept (\(\beta_{0}\)) and the coefficient for vo2_rel are -15.47 and 0.22, respectively. We can substitute these into the logistic regression equation (see Section 2) as follows:
\[P(Cold)=\frac{e^{\beta_{0}+\beta_1VO2_{rel}}}{1+e^{\beta_{0}+\beta_1VO2_{rel}}}\]
\[P(Cold)=\frac{e^{-15.47+0.22VO2_{rel}}}{1+e^{-15.47+0.22VO2_{rel}}}\]
We can interpret the coefficient on vo2_rel by exponentiating the coefficient value, \(e^{0.22}=1.25\) indicating that the odds of having a Cold increase by 25% for each additional increase in vo2_rel. We can also use the tab_model() function to quickly exponentiate the coefficients and confidence intervals:
sjPlot::tab_model(model1)| Cold | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.00 | 0.00 – 0.01 | 0.020 |
| vo2 rel | 1.25 | 1.06 – 1.58 | 0.025 |
| Observations | 25 | ||
| R2 Tjur | 0.269 | ||
4.4.2 Categorical predictor
Suppose we wanted to build a model with the categorical variable, group. In R, this would be written as:
model2 <- glm(Cold ~ group,
family = binomial,
data = Illness_post)And using the sjPlot package, we quickly obtain the exponentiated coefficients and confidence intervals:
sjPlot::tab_model(model2)| Cold | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.60 | 0.12 – 2.45 | 0.484 |
| group [4x(8x40/20s)] | 0.48 | 0.05 – 3.94 | 0.494 |
| group [4x8min] | 1.00 | 0.13 – 7.98 | 1.000 |
| Observations | 25 | ||
| R2 Tjur | 0.025 | ||
We can interpret these results as:
- The odds of having a cold are 52% lower for those in the 4x(8x40/20s) group compared to those in the 4x(12x40/20s) group, albeit this is not statistically significant.
- The odds of having a cold are approximately the same for those in the 4x(4x8min) group compared to those in the 4x(12x40/20s) group, albeit this is not statistically significant.
Note that our sample size (n = 25) is quite small, so there is a possibility that we just don’t have the statistical power to detect these effects.
4.4.3 Multiple predictors
Similar to linear regression, we can construct our logistic regression models to include multiple predictors:
model3 <- glm(Cold ~ vo2_rel + group,
family = binomial,
data = Illness_post)sjPlot::tab_model(model3)| Cold | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.00 | 0.00 – 0.01 | 0.024 |
| vo2 rel | 1.26 | 1.06 – 1.65 | 0.028 |
| group [4x(8x40/20s)] | 0.35 | 0.02 – 4.29 | 0.425 |
| group [4x8min] | 0.90 | 0.07 – 11.17 | 0.933 |
| Observations | 25 | ||
| R2 Tjur | 0.290 | ||
We can interpret these results as:
- The odds of having a Cold increase by 26% for each additional unit increase in
vo2_rel(95% CI = 1.06 to 1.65, p = .028), when controlling for group. - The odds of having a cold are 65% lower for those in the 4x(8x40/20s) group compared to those in the 4x(12x40/20s) group, albeit this is not statistically significant, when controlling for
vo2_rel. - The odds of having a cold are 10% lower for those in the 4x(4x8min) group compared to those in the 4x(12x40/20s) group, albeit this is not statistically significant, when controlling for
vo2_rel.
4.4.4 Interaction Effects
Earlier, when we plotted the empirical logits for vo2_rel, for each cycling group, we observed a potential interaction effect. Thus, we can attempt to add an interaction term to our model:
model4 <- glm(Cold ~ vo2_rel + group + vo2_rel*group,
family = binomial,
data = Illness_post)sjPlot::tab_model(model4)| Cold | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.00 | NA – 0.00 | 0.313 |
| vo2 rel | 2.35 | 1.13 – NA | 0.314 |
| group [4x(8x40/20s)] | 237836171748565581824.00 | 0.00 – NA | 0.417 |
| group [4x8min] | 50044898614373277696.00 | 0.00 – NA | 0.432 |
| vo2 rel × group [4x(8x40/20s)] |
0.49 | NA – 1.17 | 0.406 |
| vo2 rel × group [4x8min] | 0.51 | NA – 1.22 | 0.429 |
| Observations | 25 | ||
| R2 Tjur | 0.360 | ||
In the displayed output, it’s evident that several odds ratios are reported with astronomical estimates, while some confidence intervals are marked as NA (not available). This phenomenon arises due to the limited size of our sample. Particularly, when we disaggregate the dependent variable across all levels of our main and interaction effects, the precision of estimates diminishes. Consequently, accurate determination of these estimates becomes challenging within the constraints of our dataset. Thus, we should revert back to model3, which did not include the interaction effect.
4.5 Evaluating our model
summary(model3)
Call:
glm(formula = Cold ~ vo2_rel + group, family = binomial, data = Illness_post)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -16.0594 7.1179 -2.256 0.0241 *
vo2_rel 0.2347 0.1065 2.204 0.0275 *
group4x(8x40/20s) -1.0488 1.3147 -0.798 0.4250
group4x8min -0.1053 1.2491 -0.084 0.9328
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 31.343 on 24 degrees of freedom
Residual deviance: 22.875 on 21 degrees of freedom
AIC: 30.875
Number of Fisher Scoring iterations: 5In Regression Modeling Strategies 1 - Linear Regression, we learnt how to use fit metrics such as R^2, BIC, RMSE, etc., to evaluate our model. For logistic regression, one approach is to use our model to compute the probability that each subject has the cold:
probabilities <- model3 %>% predict(Illness_post, type = "response")
head(probabilities) 1 2 3 4 5 6
0.1550538 0.6730356 0.4344277 0.8892623 0.1847837 0.4058471 In the output above (which shows the first 6 subjects), probabilities of having a cold were computed based upon subject’s vo2_rel measurements and cycling group allocation. For example, subject 1 only has a 15.5% chance, whereas subject 4 has an 88.9% chance, of having a cold based upon the model.
Using an arbitrary threshold (default: 0.50), we can classify the probabilities into 1 = ‘cold’ or 0 = ‘no cold’:
predicted.classes <- ifelse(probabilities > .5, 1, 0)
cbind(probabilities, predicted.classes) |> head() probabilities predicted.classes
1 0.1550538 0
2 0.6730356 1
3 0.4344277 0
4 0.8892623 1
5 0.1847837 0
6 0.4058471 0From the output above (whih shows the first 6 subjects), we can see that using a threshold of 0.50, subject 2 (probability = 0.67) and subject 4 (probability = 0.89) would be predicted to be having a cold. From these 6 subjects, the remaining 4 subjects have probabilities less than the threshold of 0.50, therefore they would be predicted to be not having a cold.
Finally, we can compare these predicted classes to whether or not the subjects actually had a cold:
library(caret)
cm <- confusionMatrix(factor(predicted.classes, levels = c(1,0)),
factor(Illness_post$Cold1, levels = c(1, 0)))
cm$table Reference
Prediction 1 0
1 3 2
0 5 15These results reveal that:
- Of the 5 subjects predicted to have the cold, 3 of them did have the cold
- Of the 20 subjects predicted to not have the cold, 15 of them did not have the cold
Based upon these figures, the overall accuracy of this model is 72%:
cm$overall[[1]][1] 0.72Now, whilst this does seem like a reasonably high accuracy rating, there are some issues that we should consider:
- We did not split the data into training and testing sets (see Lesson 1). Due to our small sample, splitting the data would make it very difficult to estimate the effects. This meant that we made predictions on the same data that was used to build the model. This likely would have resulted in overfitting the model, explaining the high accuracy rating.
- We chose an arbitrary value of 0.50 for the threshold to predict classes. Choosing a threshold without considering the specific context or the costs associated with false positives and false negatives may not be optimal. Different threshold values could lead to varying trade-offs between sensitivity and specificity, which are important considerations in binary classification tasks. Methods such as Receiver Operating Characteristic (ROC) curve analysis to calculate Area Under the Curve (AUC) can help with this.
5 Conclusion and Reflection
In conclusion, this guide has provided an overview of regression modeling techniques suitable for binary variables, beginning with the assumptions underlying such models. We explored logistic regression models different types of predictors: continuous, categorical, continuous an categorical, and interactions. We learnt how to make predictions with our model to assess the accuracy of classification.
Throughout our discussion, we applied these regression techniques to a practical example of predicting whether or not someone has he cold, using two predictor variables. In our example we saw the limitations of a small data set, and it’s impact on interpretability. Nonetheless, this lesson provides a guide for you to be able to implement your own logistic regression models.
6 Knowledge Spot Check
What type of outcome variable is typically used in logistic regression?
- Continuous
- Binary
- Count
- Ordinal
- Binary
Logistic regression is commonly used for binary outcome variables, where the dependent variable has two possible outcomes, such as success/failure, yes/no, or presence/absence.
What is the link function commonly used in logistic regression?
- Identity function
- Logit function
- Exponential function
- Log function
- Logit function
The logit function, also known as the logistic or log-odds function, is the link function in logistic regression. It models the log-odds of the probability of the event occurring as a linear function of the predictor variables.
In logistic regression, what does an odds ratio (OR) greater than 1 indicate?
- The event is less likely to occur as the predictor variable increases.
- The event probability decreases with the predictor variable.
- The event is more likely to occur as the predictor variable increases.
- There is no relationship between the predictor variable and the event probability.
- The event is more likely to occur as the predictor variable increases.
An odds ratio greater than 1 indicates that the odds of the event occurring increase as the predictor variable increases, holding all other variables constant.
What does the assumption of linearity in logistic regression refer to?
- The predictors must have a linear relationship with the outcome variable.
- The residuals of the model must be linear.
- The outcome variable must be linearly distributed.
- The predictors must have a linear relationship with the log-odds of the outcome variable.
- The predictors must have a linear relationship with the log-odds of the outcome variable.
In logistic regression, the assumption of linearity pertains to the relationship between the predictor variables and the log-odds (logit) of the outcome, not the outcome variable itself.
Which of the following best describes why multicollinearity is problematic in logistic regression?
- It increases the model’s accuracy.
- It makes it difficult to isolate the effect of individual predictors on the outcome.
- It causes the outcome variable to be skewed.
- It violates the assumption of binary outcomes.
- It makes it difficult to isolate the effect of individual predictors on the outcome.
Multicollinearity arises when two or more predictor variables are highly correlated, leading to instability in coefficient estimates and making it difficult to determine the unique contribution of each variable.